Difference between revisions of "Talk:Addon:DNASegmentMapGramplet"
GaryGriffin (talk | contribs) |
GaryGriffin (talk | contribs) (Future version of documentation - pending PR merge) |
||
| Line 1: | Line 1: | ||
| − | [[ | + | {{languages|Addon:DNASegmentMapGramplet}} |
| + | {{Third-party plugin}} | ||
| + | [[File:Addon-DNA-SegmentMap-with-Tooltip2.png|thumb|450px|right|[[Gramps_{{man version}}_Wiki_Manual_-_Gramplets#The_split-screen_Sidebar_.26_Bottombar|Undocked]] {{man label|DNA Segment Map Gramplet}} with the HighContrast [[Addon:Themes|theme]]]] | ||
| + | The {{man label|DNA Segment Map Gramplet}} shows a graph. | ||
| − | + | =Usage= | |
| + | The purpose of this gramplet is to view DNA segment data for the active user and a set of associated users. Once the user has done an autosomal DNA test and uploaded their data to one of the vendors (GEDmatch or FamilyTreeDNA, for example), the vendor can calculate the shared DNA segments with others in their system. These list of people with their shared segments are input to this gramplet for visualization. | ||
| − | The | + | Each person with shared segments will be a separate Association. The Notes in the Association should contain the shared segment info as calculated by the vendor. |
| − | The | + | There are two views for this gramplet. The default view is all of the chromosomes with all of the segments, painted in the order of the Association. The second view is a detailed view of a single chromosome, with each associated person having a separate row. The user clicks on the chromosome label (y-axis) to switch to the detailed view and clicks on the background to return to the default view. |
| − | + | You can install the {{man label|DNA Segment Map Gramplet}} on the bottombar of one of the people or relationship category list views. | |
| − | [[ | + | ==Create a DNA Association== |
| + | To specify shared DNA segments between 2 people, | ||
| + | * Create an Association for one person (Person A) with another Person (Person B) of the type DNA. | ||
| + | * Create a Note in the Association or attached to a Citation in the Association with the shared DNA segment data. | ||
| + | ** The format of the Note is a comma separated list or a tab separated list in the order: <code>[https://wikipedia.org/wiki/Chromosome#Human_chromosomes Chromosome Number], Start Segment, End Segment, shared length in [[Genealogy_Glossary#centimorgan|centiMorgans]] (cMs), [https://wikipedia.org/wiki/Single-nucleotide_polymorphism SNP] (optional), M or P or U (unknown) to override the Maternal Paternal chromosome as determined by the closest genetic connection in the tree (optional). </code>. | ||
| + | ** e.g.: <code>3,56950055,64247327,10.9,1404</code> Which means; Chromosome Number: 3, Start Segment: 56950055, End Segment: 64247327, shared length in cMs: 10.9, matching SNPs: 1404</code> | ||
| + | ** Valid entries for each are: | ||
| − | - | + | ;Chromosome Number: number between 1-22 or X |
| − | + | ;Start Segment: The starting number for the segment location. | |
| − | + | ;End Segment: The ending number for the segment location. | |
| − | + | ;Shared length in cMs: The Genetic Distance (otherwise known as the number of centiMorgans) in the segment. | |
| − | + | ;SNP: optional field of the matching SNPs (Single Nucleotide Polymorphism) in the segment. | |
| + | ;M/P flag: optional field to override the Maternal or Paternal or Unknown chromosome. Valid entries are M or P or U. Any other data is ignored | ||
| + | |||
| + | {{-}} | ||
| + | ===Getting the chromosome data === | ||
| + | Sites like [https://www.gedmatch.com/login1.php GEDmatch] make this shared chromosome data available. Direct copy from the GEDmatch results page (with header and tab separators) will work. There can be additional Associations between Person A and Person C (et cetera) as known. | ||
| + | |||
| + | [https://dnapainter.com/help/matchdata DNApainter] provides a description of how to get the chromosome data from many of the common sites. | ||
| + | |||
| + | ===Legend=== | ||
| + | [[File:Addon-DNA-SegmentMap-legend.png|thumb|338px|right|[ {{man label|Legend with rollover tooltip]] | ||
| + | * For each Chromosome: the top portion is the Paternal side and the bottom portion is the Maternal side. | ||
| + | * The chromosome segment side (Paternal or Maternal) is determined from the Most Recent Common Ancestor. If there is no common ancestor, both sides are used. | ||
| + | * The color code for each associated person in the DNA segment map is consistent but not user-specified. The first Association will always be the same color. | ||
| + | |||
| + | ===Navigation=== | ||
| + | * The Legend on the right side lists each associated person who has a mapped segment. Hovering over the legend items will show a tooltip for possible action. Primary button click will change the active person. Secondary button click will open the Person Editor for the associated person. | ||
| + | * Hovering over the Y-axis chromosome labels will show a tooltip for possible action. Primary button click will switch view to single chromosome of the label clicked. To return to full view, click on the background in the single chromosome view. | ||
| + | * Hovering over a segment provides detail on the segments at that location. If there are multiple segments overlapping, all will have details. | ||
| + | |||
| + | ===Configuration=== | ||
| + | The config file for this gramplet has the following options. Remove the comment (double semi-colon) and edit as needed. Changes are not reflected until the next time the gramps is started. | ||
| + | |||
| + | <pre> | ||
| + | [map] | ||
| + | ;;chromosome-build=37 | ||
| + | ;;legend-single-chromosome-y-offset=25 | ||
| + | ;;legend-swatch-offset-y=0 | ||
| + | ;;maternal-background=(0.833, 0.845, 0.92) | ||
| + | ;;paternal-background=(0.926, 0.825, 0.92) | ||
| + | ;;show_associate_id=1 | ||
| + | </pre> | ||
| + | |||
| + | * Chromosome Build: choose the specific build for the chromosomes. Options are 36, 37, 38. | ||
| + | * Legend Single Chromosome Y offset: Adjust the height of the legend for the single chromosome view. This may be needed if more than 12 people share the chromosome. | ||
| + | * Legend Swatch Offset: Offset for the color swatch in the legend. Should be 5 for Windows systems. 0 works for Linux and Mac | ||
| + | * Maternal Background Color: RGB values for the background of the Maternal Chromosome | ||
| + | * Paternal Background Color: RGB values for the background of the Paternal Chromosome | ||
| + | * Show Associate ID: To remove the ID on the legend and tooltip, set to 0. Otherwise leave as 1 to print the Associate ID. | ||
| + | |||
| + | |||
| + | ===DNA Example data=== | ||
| + | To reproduce the illustration with the [[Example.gramps]] dataset, create two records in the Associations tab in the Person Editor for Luther Robinson(I0656). | ||
| + | |||
| + | The first record is DNA type, adding an association with Robert F. Garner (I1123).<br /> | ||
| + | The Note under this association contains the following text: | ||
| + | |||
| + | Chromosome,Start Location,End Location,Centimorgans,Matching SNPs,Name,Match Name | ||
| + | 3,56950055,64247327,10.9,375,Luther Robinson, Robert F. Garner | ||
| + | 11,25878681,35508918,9.9,396 | ||
| + | 12,129481599,133491098,12.4,304 | ||
| + | 15,35444614,64710827,33.3,1212 | ||
| + | 1,48053426,68837810,24.6,3413 | ||
| + | 1,72956037,87857969,13.4,2035 | ||
| + | 3,69656569,74563488,9.0,974 | ||
| + | 6,6179882,15400114,18.5,1994 | ||
| + | |||
| + | |||
| + | The second record is also DNA type, adding an association with Maude Garner (I0651).<br /> | ||
| + | The Note under this association contains the following text: | ||
| + | |||
| + | 1,30578594,38686908,11.2,334 | ||
| + | 1,236520701,249210707,29.7,685 | ||
| + | 3,14446545,24339734,12.3,458 | ||
| + | 3,128688499,140766208,11.4,447 | ||
| + | 4,76585823,114118650,33.7,1317 | ||
| + | 4,163973796,190915650,49.1,1422 | ||
| + | 6,4737179,9181572,10.3,279 | ||
| + | 6,39128976,49586285,15.8,510 | ||
| + | 6,150564916,156389148,10.2,415 | ||
| + | 7,18915133,37547290,25.8,1038 | ||
| + | 7,93557588,116296896,20.2,821 | ||
| + | 7,141636563,156148608,30.9,787 | ||
| + | 8,2808265,6919748,10.7,436 | ||
| + | 8,12568161,42652859,38.7,1556 | ||
| + | 8,49039681,71454529,20.3,742 | ||
| + | 8,71990280,99554231,21.9,917 | ||
| + | 9,78958599,122204804,55.7,2014 | ||
| + | 10,5608202,10769007,10.4,333 | ||
| + | 10,19365648,38434090,19.6,775 | ||
| + | 11,26722523,37020611,11.8,447 | ||
| + | 12,66412457,94422155,24.4,1035 | ||
| + | 13,19234747,23899627,8.7,270 | ||
| + | 13,74422984,91183468,14.2,506 | ||
| + | 14,23902753,33048583,15.5,392 | ||
| + | 14,88816167,106020366,37.8,947 | ||
| + | 15,23727655,27246462,8.2,229 | ||
| + | 16,22836249,32137965,12.6,413 | ||
| + | 16,46644903,54620503,11.7,360 | ||
| + | 17,13905,6613192,18.9,419 | ||
| + | 17,25567080,44187492,19.7,694 | ||
| + | 17,44790203,72115774,39.7,1223 | ||
| + | 18,18714991,47726830,27.9,1071 | ||
| + | 18,69454453,77894844,23.5,481 | ||
| + | 19,1993444,11174625,28.1,567 | ||
| + | 19,54545531,59087479,12.7,335 | ||
| + | 20,9879166,26225145,24.3,788 | ||
| + | 20,30221104,43975451,13.5,489 | ||
| + | 21,14670124,18743733,10.8,201 | ||
| + | |||
| + | {{man tip|DNA match row data must be in a specific CSV order|The data may include a header but the Gramplet does not require one. Headers are to make the rows more readable by humans. The rows do not need to be sequentially ordered by the chromosome identifier (1 to 22 and X).}} | ||
| + | |||
| + | [[File:Addon-DNA-GEDmatch.png|thumb|600px|right]] | ||
| + | Sample GEDmatch output (see screenshot) that can be cut and paste into the Note. The fields are the same - Chromosome, Start, End, cM, and SNPs. This can be cut/paste from the GEDmatch output directly into the Note for the Association. The header line will be ignored. | ||
| + | {{-}} | ||
| + | === Example === | ||
| + | [[File:Addon-DNA-Note-Example.png|thumb|600px|right]] | ||
| + | Create an Association of type DNA as described in the [[Gramps_{{man version}}_Wiki_Manual_-_Entering_and_editing_data:_detailed_-_part_1#Associations|Association]] page to Person A. Add a Note with the DNA shared segment data. Set the Note private if you do not want the data printed in reports. | ||
| + | {{-}} | ||
| + | [[File:Addon-DNA-Association-Example.png|thumb|600px|right]] | ||
| + | Save the Association. | ||
| + | {{-}} | ||
| + | [[File:Addon-DNA-Associations-Example.png|thumb|600px|right]] | ||
| + | Add more associations as known. Each would be associated to a different person and have a different Note. Since the Associations are drawn in order, it is generally better to have them in order of closest relative to furthest relative to avoid obscuring a distant relative (smaller segment) by a close relative (larger segment). Use the up-arrow and down-arrow to change the order of the Association. | ||
| + | {{-}} | ||
| + | [[File:Addon-DNA-SegmentMap2.png|thumb|600px|right]] | ||
| + | Add the DNA gramplet to the Person view. Select the DNA tab. The segment map will be color coded by associated person. For each Chromosome the top portion is the P (Paternal) side and the M (Maternal) side is the bottom portion. If the chromosome segment side (Paternal or Maternal) is unknown, the segment will cover both the top and bottom portions of the chromosome and be 50% transparent. | ||
| + | {{-}} | ||
| + | [[File:Addon-DNA-SegmentMap-with-Tooltip2.png|thumb|600px|right]] | ||
| + | Hovering the cursor over a known segment will pop up the name of the associated person and the length (in cMs) of the shared segment. | ||
| + | {{-}} | ||
| + | [[File:Addon-DNA-SegmentMap-Single.png|thumb|600px|right]] | ||
| + | Clicking on the Y-axis label for the 6th chromosome, a detailed view is shown. Click on the background to return to the complete view. | ||
| + | {{-}} | ||
| + | |||
| + | == Reference Info == | ||
| + | === Untested Areas === | ||
| + | There are areas in the DNA test that are generally not tested as they are not reliable indicators of a match. If you want to visualize these areas, create a dummy person and add an association with the following DNA segments. | ||
| + | |||
| + | <pre> | ||
| + | 13,1,19020094,0,0 | ||
| + | 14,1,19067948,0,0 | ||
| + | 15,1,20004965,0,0 | ||
| + | 21,1,9922017,0,0 | ||
| + | 22,1,16055121,0,0 | ||
| + | </pre> | ||
| + | |||
| + | === Centromere Areas === | ||
| + | Machines have difficulties to read the area around the centromere. There are less SNPs to read, therefore there is higher probability of false positive matches. For example, if you have a match exactly around the centromere, then it is most probably a false positive match. If you want to visualize these areas, create a dummy person and add an association with the following DNA segments. | ||
<pre> | <pre> | ||
| Line 48: | Line 196: | ||
</pre> | </pre> | ||
| − | + | = To Do = | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | = Issues = | |
| − | * | + | * If the Chromosome Number is not in the range (1, 2, ..., 22, X) it is ignored. |
| − | * | + | * If there are multiple paths to a common ancestor, the closest found is used. |
| − | * | + | * To create a segment map for Person A, you need to add associations to Person A. There is no reciprocal relationship for Person B - that is, there is no segment map for Person B, only for Person A. You can execute the [[Addon:SyncAssociation]] addon to create any missing reciprocal relationships. |
| − | * | + | * Color code for each associated person in the map is consistent but not user-specified. The first Association will always be the same color. |
| − | + | * If there are overlapping segments within a maternal/paternal view of a chromosome, only the front (last drawn) will be pickable. The tooltip will still provide the details of the hidden segments. Changing the order of Associations (using the up-arrow and down-arrow) to have the closer relatives before further relatives will fix this. | |
| − | |||
| − | + | = See also = | |
| + | * DNA Segment Map gramplet #550 (GitHub pre-release [https://github.com/gramps-project/addons-source/pull/550]) | ||
| + | * [[Addon:SyncAssociation]] - For every Association, if there is not a reciprocal Association then it will be created. That is, where Person John Smith has a DNA Association (with an Association Note) to Person Jane Jones, then if Jane Jones does not have a corresponding DNA Association to John Smith, it will be created which shares the Note. | ||
| + | * [[Add Types to the SyncAssociation Gramplet]] | ||
| + | * [https://gramps.discourse.group/t/updated-dna-gramplet-ready-for-review/807 Discourse forum discussion on the DNAgramplet] | ||
| + | * [[Gramps_{{man version}}_Wiki_Manual_-_Entering_and_editing_data:_detailed_-_part_1#Associations|Introduction to Associations in Gramps]] | ||
| + | * [[Roles, Relationships & Associations]] | ||
| + | * [[Genetics]] | ||
| − | + | [[Category:Plugins]] | |
| + | [[Category:Addons]] | ||
Revision as of 18:59, 5 November 2022
This is a Third-party Addon. Please use carefully on data that is backed up, and help make it better by reporting any comments or problems to the author, or issues to the bug tracker |
The DNA Segment Map Gramplet shows a graph.
Contents
Usage
The purpose of this gramplet is to view DNA segment data for the active user and a set of associated users. Once the user has done an autosomal DNA test and uploaded their data to one of the vendors (GEDmatch or FamilyTreeDNA, for example), the vendor can calculate the shared DNA segments with others in their system. These list of people with their shared segments are input to this gramplet for visualization.
Each person with shared segments will be a separate Association. The Notes in the Association should contain the shared segment info as calculated by the vendor.
There are two views for this gramplet. The default view is all of the chromosomes with all of the segments, painted in the order of the Association. The second view is a detailed view of a single chromosome, with each associated person having a separate row. The user clicks on the chromosome label (y-axis) to switch to the detailed view and clicks on the background to return to the default view.
You can install the DNA Segment Map Gramplet on the bottombar of one of the people or relationship category list views.
Create a DNA Association
To specify shared DNA segments between 2 people,
- Create an Association for one person (Person A) with another Person (Person B) of the type DNA.
- Create a Note in the Association or attached to a Citation in the Association with the shared DNA segment data.
- The format of the Note is a comma separated list or a tab separated list in the order:
Chromosome Number, Start Segment, End Segment, shared length in centiMorgans (cMs), SNP (optional), M or P or U (unknown) to override the Maternal Paternal chromosome as determined by the closest genetic connection in the tree (optional).. - e.g.:
3,56950055,64247327,10.9,1404Which means; Chromosome Number: 3, Start Segment: 56950055, End Segment: 64247327, shared length in cMs: 10.9, matching SNPs: 1404 - Valid entries for each are:
- The format of the Note is a comma separated list or a tab separated list in the order:
- Chromosome Number
- number between 1-22 or X
- Start Segment
- The starting number for the segment location.
- End Segment
- The ending number for the segment location.
- Shared length in cMs
- The Genetic Distance (otherwise known as the number of centiMorgans) in the segment.
- SNP
- optional field of the matching SNPs (Single Nucleotide Polymorphism) in the segment.
- M/P flag
- optional field to override the Maternal or Paternal or Unknown chromosome. Valid entries are M or P or U. Any other data is ignored
Getting the chromosome data
Sites like GEDmatch make this shared chromosome data available. Direct copy from the GEDmatch results page (with header and tab separators) will work. There can be additional Associations between Person A and Person C (et cetera) as known.
DNApainter provides a description of how to get the chromosome data from many of the common sites.
Legend
- For each Chromosome: the top portion is the Paternal side and the bottom portion is the Maternal side.
- The chromosome segment side (Paternal or Maternal) is determined from the Most Recent Common Ancestor. If there is no common ancestor, both sides are used.
- The color code for each associated person in the DNA segment map is consistent but not user-specified. The first Association will always be the same color.
- The Legend on the right side lists each associated person who has a mapped segment. Hovering over the legend items will show a tooltip for possible action. Primary button click will change the active person. Secondary button click will open the Person Editor for the associated person.
- Hovering over the Y-axis chromosome labels will show a tooltip for possible action. Primary button click will switch view to single chromosome of the label clicked. To return to full view, click on the background in the single chromosome view.
- Hovering over a segment provides detail on the segments at that location. If there are multiple segments overlapping, all will have details.
Configuration
The config file for this gramplet has the following options. Remove the comment (double semi-colon) and edit as needed. Changes are not reflected until the next time the gramps is started.
[map] ;;chromosome-build=37 ;;legend-single-chromosome-y-offset=25 ;;legend-swatch-offset-y=0 ;;maternal-background=(0.833, 0.845, 0.92) ;;paternal-background=(0.926, 0.825, 0.92) ;;show_associate_id=1
- Chromosome Build: choose the specific build for the chromosomes. Options are 36, 37, 38.
- Legend Single Chromosome Y offset: Adjust the height of the legend for the single chromosome view. This may be needed if more than 12 people share the chromosome.
- Legend Swatch Offset: Offset for the color swatch in the legend. Should be 5 for Windows systems. 0 works for Linux and Mac
- Maternal Background Color: RGB values for the background of the Maternal Chromosome
- Paternal Background Color: RGB values for the background of the Paternal Chromosome
- Show Associate ID: To remove the ID on the legend and tooltip, set to 0. Otherwise leave as 1 to print the Associate ID.
DNA Example data
To reproduce the illustration with the Example.gramps dataset, create two records in the Associations tab in the Person Editor for Luther Robinson(I0656).
The first record is DNA type, adding an association with Robert F. Garner (I1123).
The Note under this association contains the following text:
Chromosome,Start Location,End Location,Centimorgans,Matching SNPs,Name,Match Name 3,56950055,64247327,10.9,375,Luther Robinson, Robert F. Garner 11,25878681,35508918,9.9,396 12,129481599,133491098,12.4,304 15,35444614,64710827,33.3,1212 1,48053426,68837810,24.6,3413 1,72956037,87857969,13.4,2035 3,69656569,74563488,9.0,974 6,6179882,15400114,18.5,1994
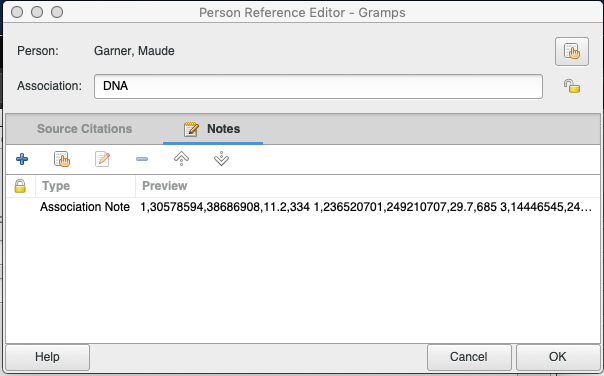
The second record is also DNA type, adding an association with Maude Garner (I0651).
The Note under this association contains the following text:
1,30578594,38686908,11.2,334 1,236520701,249210707,29.7,685 3,14446545,24339734,12.3,458 3,128688499,140766208,11.4,447 4,76585823,114118650,33.7,1317 4,163973796,190915650,49.1,1422 6,4737179,9181572,10.3,279 6,39128976,49586285,15.8,510 6,150564916,156389148,10.2,415 7,18915133,37547290,25.8,1038 7,93557588,116296896,20.2,821 7,141636563,156148608,30.9,787 8,2808265,6919748,10.7,436 8,12568161,42652859,38.7,1556 8,49039681,71454529,20.3,742 8,71990280,99554231,21.9,917 9,78958599,122204804,55.7,2014 10,5608202,10769007,10.4,333 10,19365648,38434090,19.6,775 11,26722523,37020611,11.8,447 12,66412457,94422155,24.4,1035 13,19234747,23899627,8.7,270 13,74422984,91183468,14.2,506 14,23902753,33048583,15.5,392 14,88816167,106020366,37.8,947 15,23727655,27246462,8.2,229 16,22836249,32137965,12.6,413 16,46644903,54620503,11.7,360 17,13905,6613192,18.9,419 17,25567080,44187492,19.7,694 17,44790203,72115774,39.7,1223 18,18714991,47726830,27.9,1071 18,69454453,77894844,23.5,481 19,1993444,11174625,28.1,567 19,54545531,59087479,12.7,335 20,9879166,26225145,24.3,788 20,30221104,43975451,13.5,489 21,14670124,18743733,10.8,201
Sample GEDmatch output (see screenshot) that can be cut and paste into the Note. The fields are the same - Chromosome, Start, End, cM, and SNPs. This can be cut/paste from the GEDmatch output directly into the Note for the Association. The header line will be ignored.
Example
Create an Association of type DNA as described in the Association page to Person A. Add a Note with the DNA shared segment data. Set the Note private if you do not want the data printed in reports.
Save the Association.
Add more associations as known. Each would be associated to a different person and have a different Note. Since the Associations are drawn in order, it is generally better to have them in order of closest relative to furthest relative to avoid obscuring a distant relative (smaller segment) by a close relative (larger segment). Use the up-arrow and down-arrow to change the order of the Association.
Add the DNA gramplet to the Person view. Select the DNA tab. The segment map will be color coded by associated person. For each Chromosome the top portion is the P (Paternal) side and the M (Maternal) side is the bottom portion. If the chromosome segment side (Paternal or Maternal) is unknown, the segment will cover both the top and bottom portions of the chromosome and be 50% transparent.
Hovering the cursor over a known segment will pop up the name of the associated person and the length (in cMs) of the shared segment.
Clicking on the Y-axis label for the 6th chromosome, a detailed view is shown. Click on the background to return to the complete view.
Reference Info
Untested Areas
There are areas in the DNA test that are generally not tested as they are not reliable indicators of a match. If you want to visualize these areas, create a dummy person and add an association with the following DNA segments.
13,1,19020094,0,0 14,1,19067948,0,0 15,1,20004965,0,0 21,1,9922017,0,0 22,1,16055121,0,0
Centromere Areas
Machines have difficulties to read the area around the centromere. There are less SNPs to read, therefore there is higher probability of false positive matches. For example, if you have a match exactly around the centromere, then it is most probably a false positive match. If you want to visualize these areas, create a dummy person and add an association with the following DNA segments.
1,121535434,124535434,0,0 2,92326171,95326171,0,0 3,90504854,93504854,0,0 4,49660117,52660117,0,0 5,46405641,49405641,0,0 6,58830166,61830166,0,0 7,58054331,61054331,0,0 8,43838887,46838887,0,0 9,47367679,50367679,0,0 10,39254935,42254935,0,0 11,51644205,54644205,0,0 12,34856694,37856694,0,0 13,16000000,19000000,0,0 14,16000000,19000000,0,0 15,17000000,20000000,0,0 16,35335801,38335801,0,0 17,22263006,25263006,0,0 18,15460898,18460898,0,0 19,24681782,27681782,0,0 20,26369569,29369569,0,0 21,11288129,14288129,0,0 22,13000000,16000000,0,0 X,58632012,61632012,0,0
To Do
Issues
- If the Chromosome Number is not in the range (1, 2, ..., 22, X) it is ignored.
- If there are multiple paths to a common ancestor, the closest found is used.
- To create a segment map for Person A, you need to add associations to Person A. There is no reciprocal relationship for Person B - that is, there is no segment map for Person B, only for Person A. You can execute the Addon:SyncAssociation addon to create any missing reciprocal relationships.
- Color code for each associated person in the map is consistent but not user-specified. The first Association will always be the same color.
- If there are overlapping segments within a maternal/paternal view of a chromosome, only the front (last drawn) will be pickable. The tooltip will still provide the details of the hidden segments. Changing the order of Associations (using the up-arrow and down-arrow) to have the closer relatives before further relatives will fix this.
See also
- DNA Segment Map gramplet #550 (GitHub pre-release [1])
- Addon:SyncAssociation - For every Association, if there is not a reciprocal Association then it will be created. That is, where Person John Smith has a DNA Association (with an Association Note) to Person Jane Jones, then if Jane Jones does not have a corresponding DNA Association to John Smith, it will be created which shares the Note.
- Add Types to the SyncAssociation Gramplet
- Discourse forum discussion on the DNAgramplet
- Introduction to Associations in Gramps
- Roles, Relationships & Associations
- Genetics